Abstract
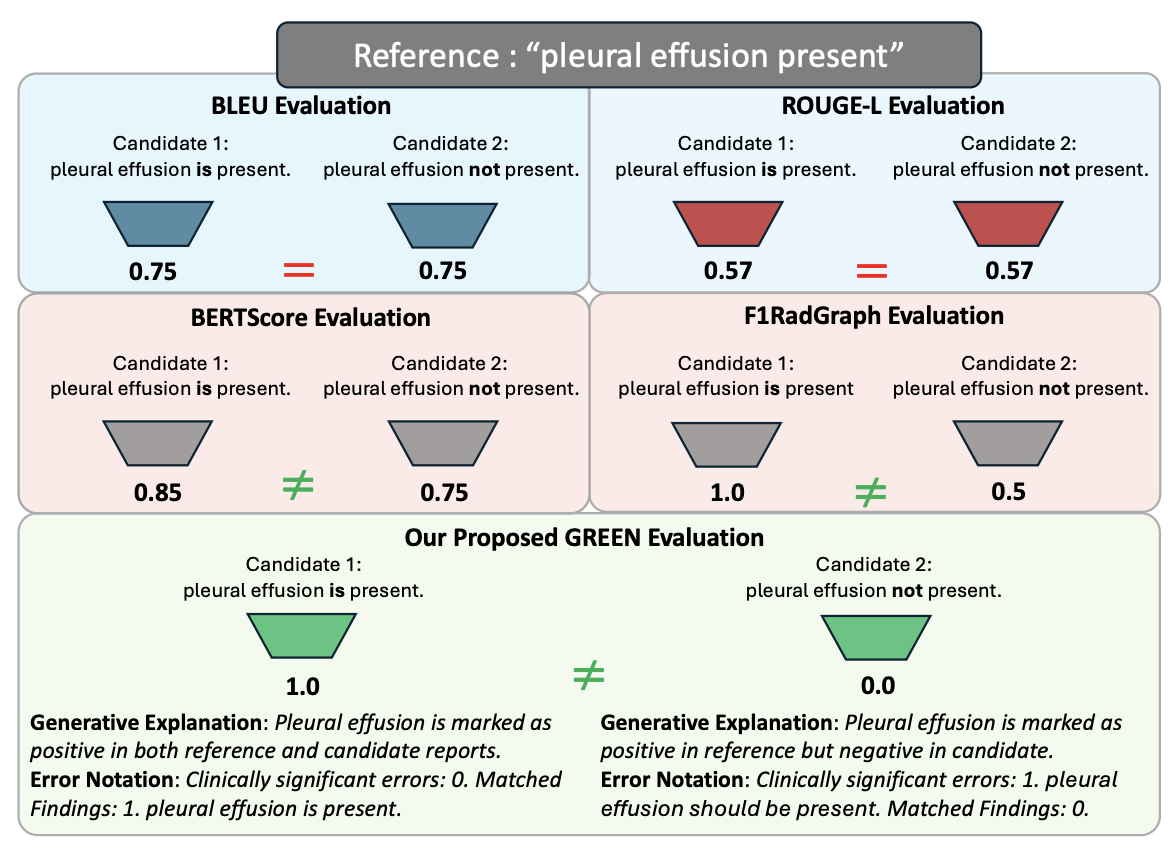
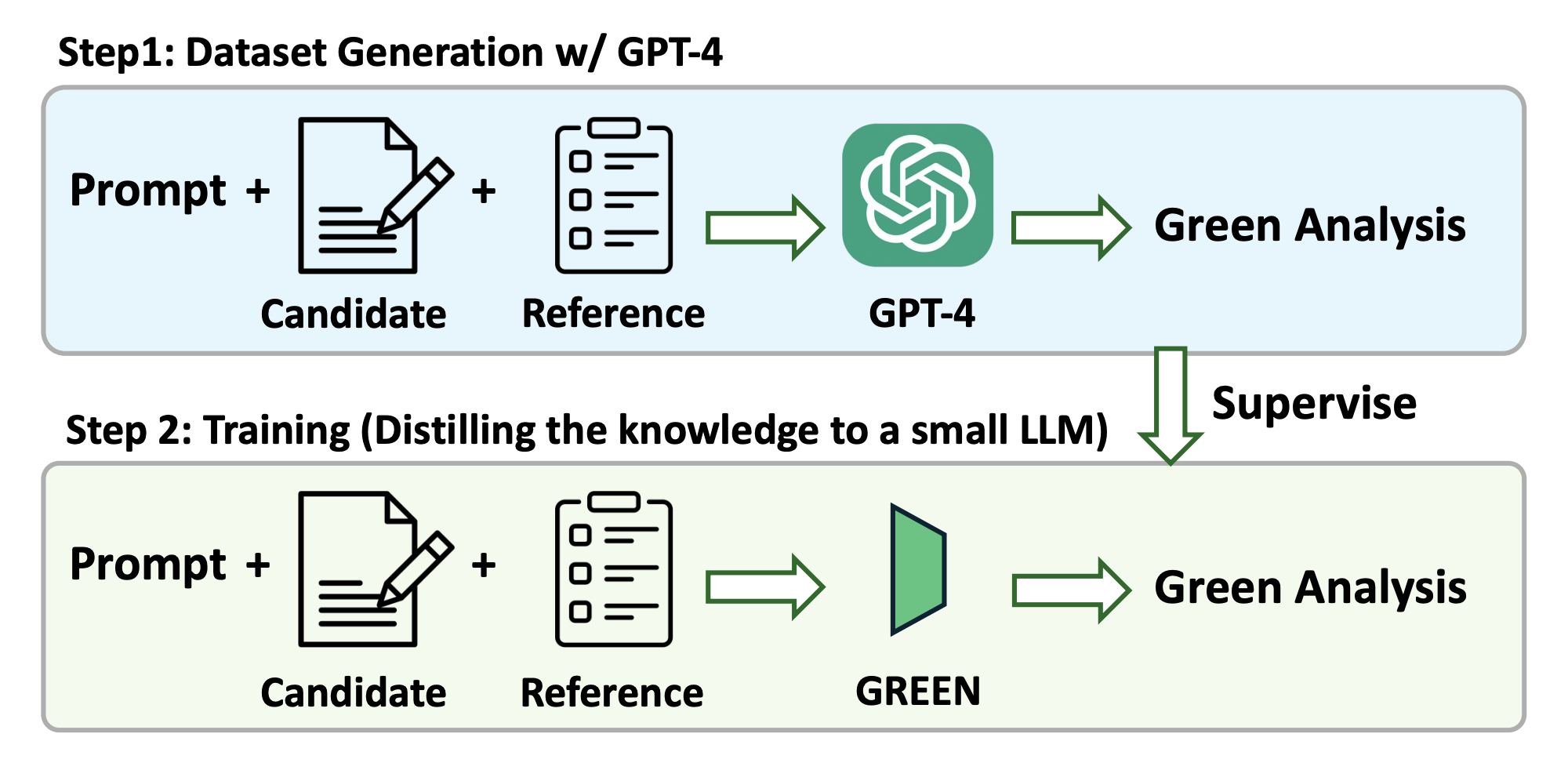
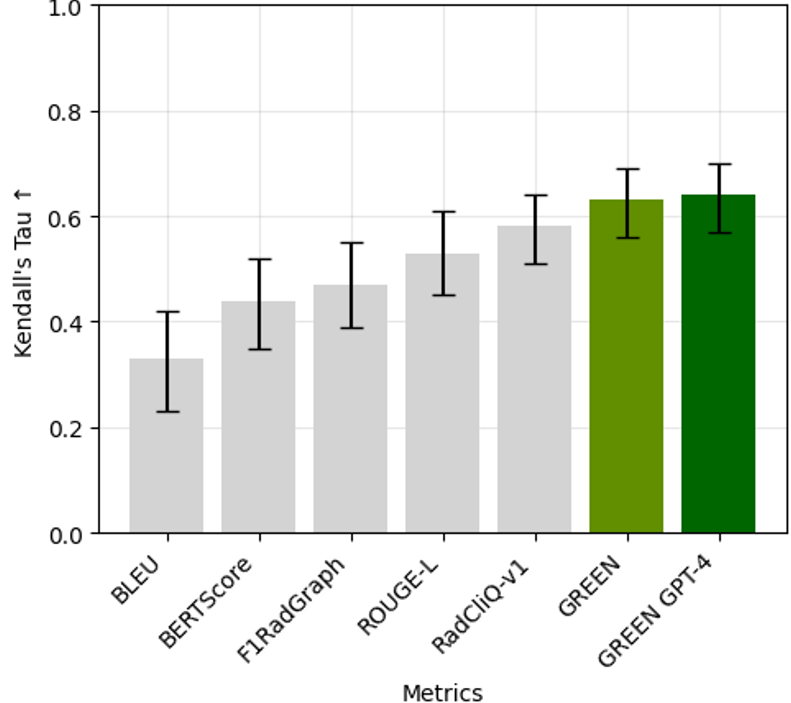
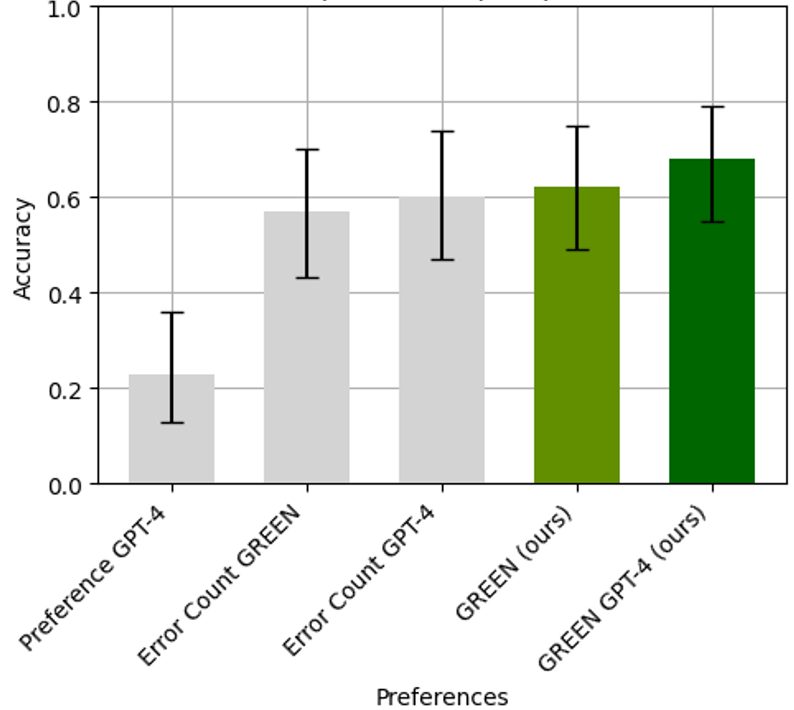
Evaluating radiology reports is a challenging problem as factual correctness is extremely important due to its medical nature. Existing automatic evaluation metrics either suffer from failing to consider factual correctness (e.g., BLEU and ROUGE) or are limited in their interpretability (e.g., F1CheXpert and F1RadGraph). In this paper, we introduce GREEN (Generative Radiology Report Evaluation and Error Notation), a radiology report generation metric that leverages the natural language understanding of language models to identify and explain clinically significant errors in candidate reports, both quantitatively and qualitatively. Compared to current metrics, GREEN offers a score aligned with expert preferences, human interpretable explanations of clinically significant errors, enabling feedback loops with end-users, and a lightweight open-source method that reaches the performance of commercial counterparts. We validate our GREEN metric by comparing it to GPT-4, as well as to the error counts of 6 experts and the preferences of 2 experts. Our method demonstrates not only a higher correlation with expert error counts but simultaneously higher alignment with expert preferences when compared to previous approaches.