Abstract
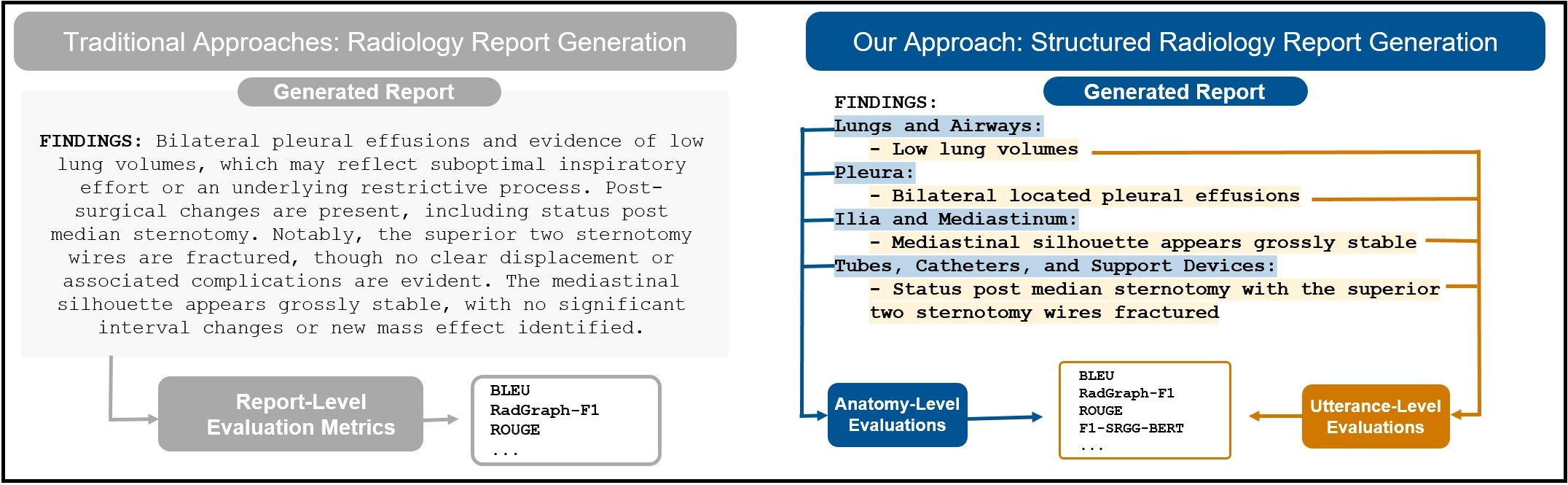
Automated radiology report generation from chest X-ray (CXR) images has the potential to improve clinical efficiency and reduce radiologists' workload. However, most datasets, including the publicly available MIMIC-CXR and CheXpert Plus, consist entirely of free-form reports, which are inherently variable and unstructured. This variability poses challenges for both generation and evaluation: existing models struggle to produce consistent, clinically meaningful reports, and standard evaluation metrics fail to capture the nuances of radiological interpretation. To address this, we introduce Structured Radiology Report Generation (SRRG), a new task that reformulates free-text radiology reports into a standardized format, ensuring clarity, consistency, and structured clinical reporting. We create a novel dataset by restructuring reports using large language models (LLMs) following strict structured reporting desiderata. Additionally, we introduce SRR-BERT, a fine-grained disease classification model trained on 55 labels, enabling more precise and clinically informed evaluation of structured reports. To assess report quality, we propose F1-SRR-BERT, a metric that leverages SRR-BERT’s hierarchical disease taxonomy to bridge the gap between free-text variability and structured clinical reporting. We validate our dataset through a reader study conducted by five board-certified radiologists and extensive benchmarking experiments.