Abstract
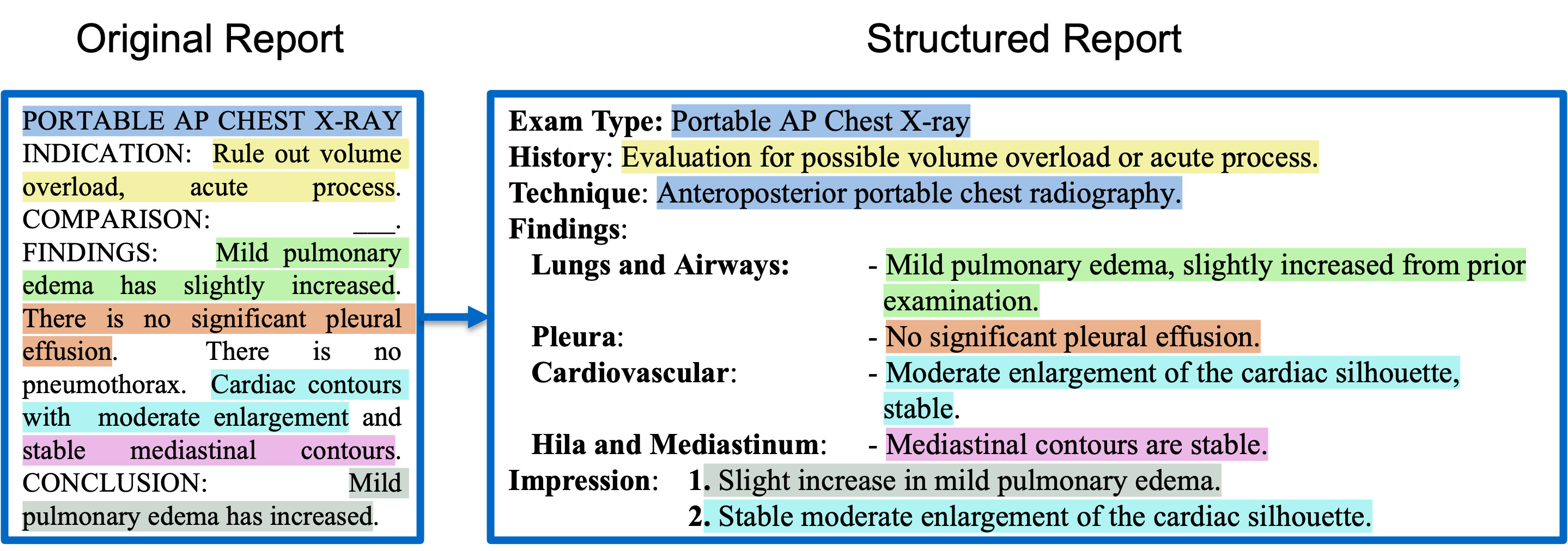
Radiology reports are critical for clinical decision-making but often lack a standardized format, limiting both human interpretability and machine learning (ML) applications. While large language models (LLMs) have shown strong capabilities in reformatting clinical text, their high computational requirements, lack of transparency, and data privacy concerns hinder practical deployment. To address these challenges, we explore lightweight encoder-decoder models (<300M parameters)—specifically T5 and BERT2BERT—for structuring radiology reports from the MIMIC-CXR and CheXpert Plus datasets. We benchmark these models against eight open-source LLMs (1B–70B parameters), adapted using prefix prompting, in-context learning (ICL), and low-rank adaptation (LoRA) finetuning. Our best-performing lightweight model outperforms all LLMs adapted using prompt-based techniques on a human-annotated test set. While some LoRA-finetuned LLMs achieve modest gains over the lightweight model on the Findings section (BLEU 6.4%, ROUGE-L 4.8%, BERTScore 3.6%, F1-RadGraph 1.1%, GREEN 3.6%, and F1-SRR-BERT 4.3%), these improvements come at the cost of substantially greater computational resources. For example, LLaMA-3-70B incurred more than 400 times the inference time, cost, and carbon emissions compared to the lightweight model. These results underscore the potential of lightweight, task-specific models as sustainable and privacy-preserving solutions for structuring clinical text in resource-constrained healthcare settings.